Liphardt Lab
Research Directions: Patterns, Energy, and Information
We're a team of physicists, engineers, and biologists. We investigate the organization and dynamics of single molecules in living cells using new optical tools. In these studies, we collect time-series data and then analyze them to find underlying regulatory and organizational principles. Our lab is located in the Shriram Center.
Beyond 'basic' biophysics, we also try to tackle broader problems in medicine. Public health and disease prevention are still significantly constrained by limited data. Current medical record systems and health AI tools are not yet optimized for rapid, private, and low cost delivery of high-quality prevention and care. However, new computational and cryptographic techniques have the potential to make health much more accessible all around the world. For example, using Secure Multiparty Computation, volunteers can allow cloud doctors to learn from symptom data and provide health recommendations, without sensitive data ever leaving a person's phone.
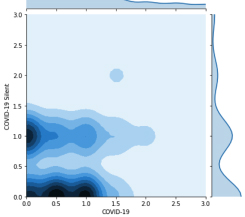
Privacy-preservivng Analytics for Digital Health
New cryptographic techniques such as Fully Homomorphic Encryption (FHE) make it possible to derive insights from encrypted data, better balancing patient privacy with the needs of public health. We recently used another technique, Secure Multiparty Computation, to investigate COVID symptoms in millions of people in 91 countries without jeopardizing their privacy.

Data analysis
We use concepts and tools from non-equilibrium statistical mechanics, machine learning, and polymer physics to model and explore biological processes. Increasingly, we use convolutional neural nets to make sense of image data. Our DeepEvolve python/keras framework for evolving CNN hyperparameters is on Github.
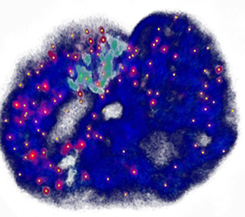
Genome-wide coordination of gene expression
Imagine you are an orchestra conductor directing a symphony. If you're good at what you do, everything will sound right. How does the genome solve the equivalent problem, except without a conductor? We use genome-edited cell lines to investigate how DNA-looping and chromatin compaction influence transcriptional regulation. The image shows a single nucleus; The DNA is blue, single RNA transcripts are red/yellow.